Working with fields¶
Nexus fields are the data holding instance in a Nexus files. On can imagine a field like a multidimensional array stored on disk. In fact the field class used in this python package is designed to behave a little like the famous numpy arrays. So they should be rather easy to use.
The simplest way to create a field would look like this
name = entry.create_field("name","str")
which creates a 1D field with a single element of type string.
The first two argument, the name of the field and its data type, are mandatory.
Like with the create_group method create_field can only create a direct
child of its parent group. The method accepts three more keyword arguments:
shape, chunk, and filter. shape determines the shape of the
field after it has been created. chunk is the chunk shape used for the
field and finally filter can be a filter instance used to compress the data
during writing. It is important to note that the create_field method will
always set a chunk size, even when none is provided by the user. This is a
requirement in order to get fields which can be grown (see next section).
This example shows field creation using all possible arguments
import pninexus.nexus as nx
path = "/:NXentry/:NXinstrument/:NXdetector"
file = nx.open_file("data.nxs",readonly=False)
detector = nx.get_object(f.root(),path)
#create a deflate filter with compression level 5
deflate = nx.deflate_filter()
deflate.rate = 5
detector.create_field("data","uint16",
shape=(0,1024,2048), #set the initial field size
chunk=(1,1024,2048), #set the chunk size
filter = deflate)
There are two things which are important here. Please note that the first
element in the shape of the field is 0. This means its total size will be 0
and no data can be stored. This is a quite reasonable approach if one does not
know how many elements (lets say image frames) will be stored in the field.
One starts with 0 and grows the field every time a new frame shall be stored.
Unlike the shape argument the chunk argument must not have 0
elements. In the example above we make the chunk size exactly one image frame
which is a reasonable choice for most applications.
Finally the above example adds a deflate filter to the field (the same filter
used for gzip) with a compression level of 5.
Field inquiry and manipulation¶
Like groups, fields have some public attributes which can be used to determine some of the properties of a field
Attribute name |
Description |
|---|---|
|
returns the name of the field |
|
returns the full nexus path of the field |
|
returns the groups parent field |
|
number of elements stored in the field |
|
data type of the elements |
|
a tuple with the number of elements along each dimension |
|
|
|
returns the name of the file the field belongs to |
Unlike numpy arrays, the shape of fields cannot be manipulated arbitrarily.
This is due to limitations of the underlying HDF5 file format.
Fortunately this is not a big issue. The most important manipulation (and
currently also the only one implemented) is to grow a field.
#create the data field for the detector
field = detector.create_field("data",...)
#main measurement loop
while measurement_running:
#execute some code retrieving the data here
field.grow(0,1) #grow field along dimension 0 by 1 element
# execute code to store data
# will be shown latter in this manual
The grow method of a field takes two positional arguments: the first is the dimension along we want to grow the field and the second is the number of elements by which we want to enlarge it.
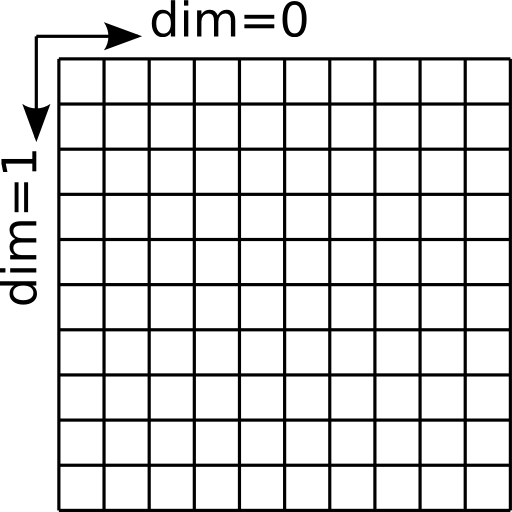
The field before growth along dimension 0¶
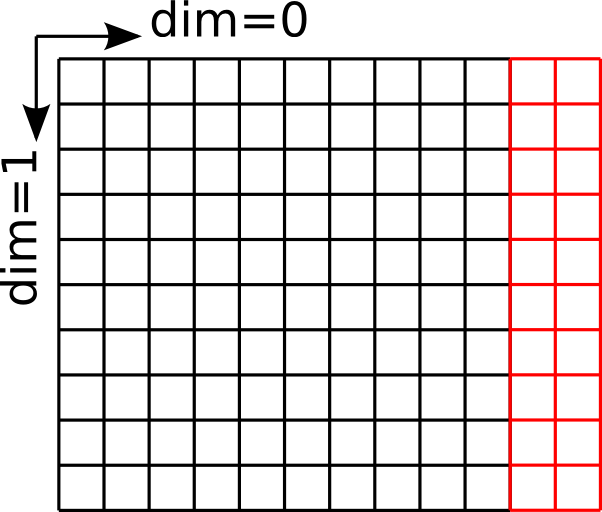
The field after extending it by two elements along dimension 0¶
Reading and writing data¶
There are two ways of how to read and write data from and to an instance of
nxfield
via its
read()andwrite()methodsor by using the
[]operator which effectively calls the__getitem__()and__setitem__()methods ofnxfield.
In the former case the entire field will be read or written while the second approach also allows partial IO which is particularly useful for large fields whose content would not fit into the main memory of the machine on which the code is running.
Using read() and write()¶
write() data writes the data to the entire field like in the
following example
from __future__ import print_function
import pninexus.nexus as nexus
f = nexus.open_file("run1.nxs",readonly=False)
root = f.root()
temperaturs = nexus.get_object(root,"/:NXentry/:NXsample/temperature")
data = read_data(...) #numpy array with temperature values
temperatures.write(data) #write all the data in a single step
For a field of size 1 the argument of write() must be a native python
object or a numpy array of size 1. If the field is a multidimensional
field of different size the argument must be a numpy array of
appropriate shape (and thus size).
The counterpart of write() is read() which retrieves all data
stored in a field
from __future__ import print_function
import pninexus.nexus as nexus
f = nexus.open_file("run1.nxs",readonly=False)
root = f.root()
#get field with positions of motor 'tt'
tt_field = nexus.get_object(root,"/:NXentry/:NXsample/:NXtransformations/tt")
# read all position values
tt = tt_field.read() #returns a numpy array with the position data
If the field is of size 1 (a scalar) a simple Python object is returned holding
the content of the field. In the case of a multidimensional field a
numpy array of appropriate type and shape is returned.
As in the h5py data can also be written from lists and nested lists.
from __future__ import print_function
import pninexus.nexus as nexus
f = nexus.create_file("run2.nxs",readonly=False)
root = f.root()
# ........ some more code to create the file ..............
eps_tensor = nexus.get_object("/:NXentry/:NXsample/strain_tensor")
eps_tensor.write([1,2,3,4,])
Partial IO with the [] operator¶
Fields follow the same indexing and broadcasting protocol as numpy
arrays. The major difference is that the data is not stored in memory but on
disk. For more details about the numpy indexing and broadcasting
protocol see the numpy reference manual.
The next code snipped shows a typical use case where a bunch of image frames is retriefed from a field by iterating over each individual image. The code should be rather self explaining
import pninexus.nexus as nx
file = nx.open_file("run_01.nxs")
root_group = file.root()
frame_path = "/:NXentry/:NXinstrument/:NXdetector/data"
#retrieve frames from the file
frames = nx.get_object(root_group,frame_path)
#iterate over the frames
for frame_index in range(frames.shape[0]):
frame_data = frames[frame_index,...]
result = do_som_work(frame_data)
Note here that the ellipse ... used for retrieving the data will make
the code independent from the actual rank of a frame. As for virtually all
other examples we assume here that the first dimension of the frame field
corresponds to the number of frames.
Writing works just the other way arround. Here we finishe the above example for
the grow() method
#create the data field for the detector
field = detector.create_field("data",...)
#main measurement loop
while measurement_running:
data = get_data(...) #retrieve data
field.grow(0,1) #grow field along dimension 0 by 1 element
field[-1,...] = data #save data in newly appended slot
Data can be written not only from numpy arrays but also from nested
lists as with h5py
field = nexus.get_object("/:NXentry/:NXinstrument/:NXdetector/data")
while measurement_running:
field.grow(0,1)
field[-1,...] = [1,2,3,4,5,6,7,8,]